import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from sklearn.metrics import log_loss
from sklearn.metrics import brier_score_loss
from sklearn.calibration import calibration_curve
from sklearn.calibration import CalibratedClassifierCV
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import GridSearchCVClassifier Calibration
Evaluating Probability Estimates
Given data with:
- \(n\), the number of samples
- \(y\), the vector of true target values
- \(y_i\) is the true label of the \(i\)th sample
- \(y_i \in \{0, 1\}\)
- \(p\), the vector of probabilities that \(y_i = 1\) given \(x_i\)
- \(p_i\) is the probability for the \(i\)th sample
- \(p_i \in [0, 1]\)
We can define two metrics to evaluate the performance of a (binary) classifier:
Log Loss
\[ \text{LogLoss}(y, p) = -\frac{1}{n} \sum_{i=1}^{n} \left[ y_i \log(p_i) + (1 - y_i) \log(1 - p_i) \right] \]
Brier Score
\[ \text{BrierScore}(y, p) = \frac{1}{n} \sum_{i=1}^{n} (y_i - p_i)^2 \]
# simulate data for binary classification
X, y = make_classification(
n_samples=5000,
n_features=4,
n_classes=2,
random_state=1,
)X[:10]array([[ 0.02803399, 0.74045307, 0.59712136, 0.31759471],
[-0.17706936, 0.34784859, 0.40240521, -0.1791632 ],
[-0.52965358, -0.71395557, -0.25371071, -1.17378147],
[-1.50936843, 3.03824685, 3.49091485, -1.50062947],
[-1.42796846, 1.43337641, 2.10561783, -1.94361204],
[ 1.95123747, -1.41096839, -2.42227469, 2.85494736],
[ 0.93718509, -1.81257251, -2.10615387, 0.95863103],
[ 0.49684818, -1.67618143, -1.71072152, 0.24817525],
[-0.80225435, -0.6585981 , -0.0330643 , -1.6241777 ],
[-0.52371064, 0.96835631, 1.13995309, -0.55188528]])y[:10]array([1, 0, 0, 1, 1, 0, 0, 0, 0, 1])np.mean(y)0.4996# train-test split the data
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.2,
random_state=42,
)# create and fit logistic regression model
model = LogisticRegression(random_state=42)
model.fit(X_train, y_train)LogisticRegression(random_state=42)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LogisticRegression(random_state=42)
# get predicted probabilities and classification
y_pred_proba = model.predict_proba(X_test)[:, 1]
y_pred = model.predict(X_test)# calculate metrics
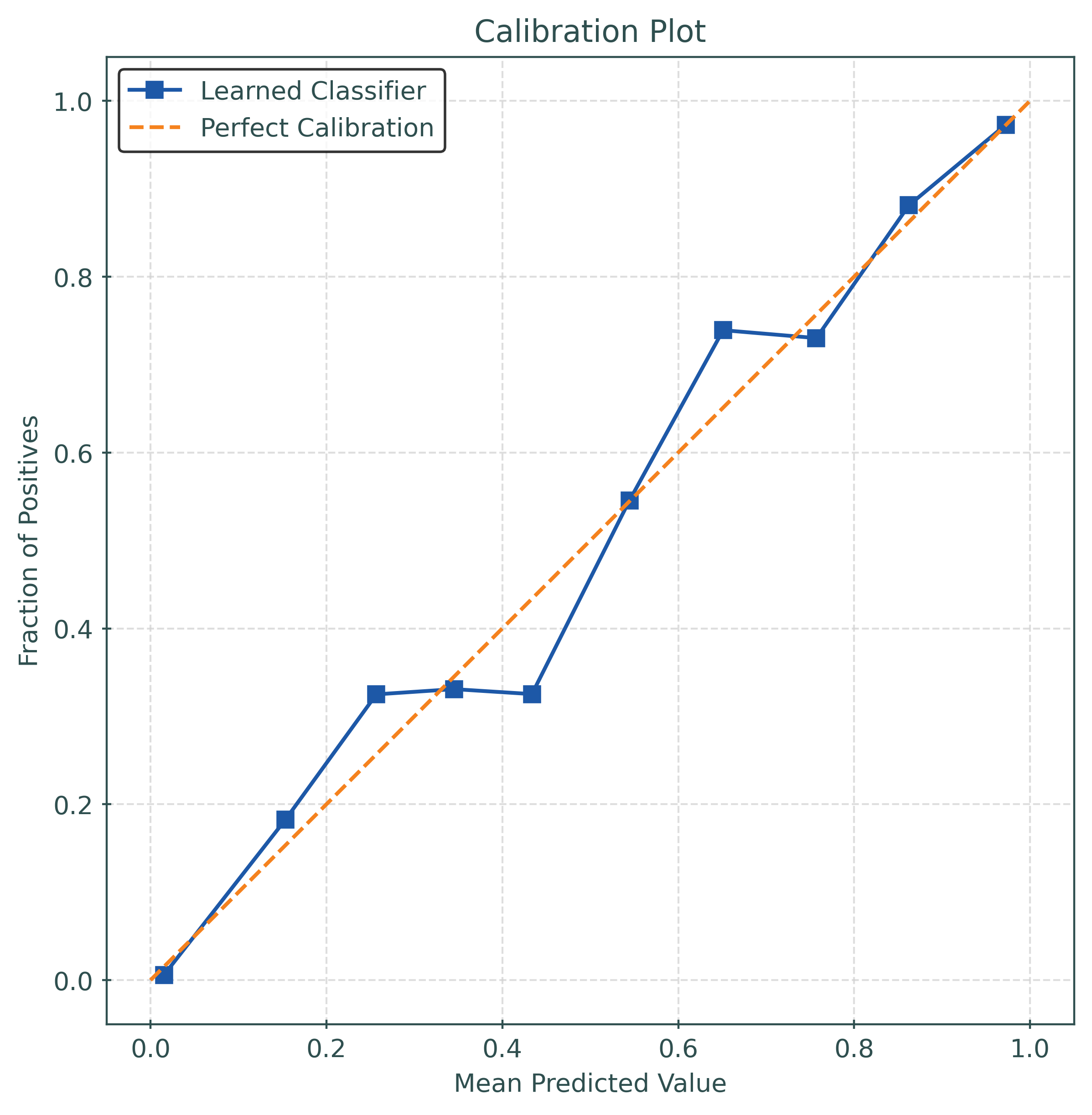
test_accuracy = accuracy_score(y_test, y_pred)
test_logloss = log_loss(y_test, y_pred_proba)
test_brier_score = brier_score_loss(y_test, y_pred_proba)
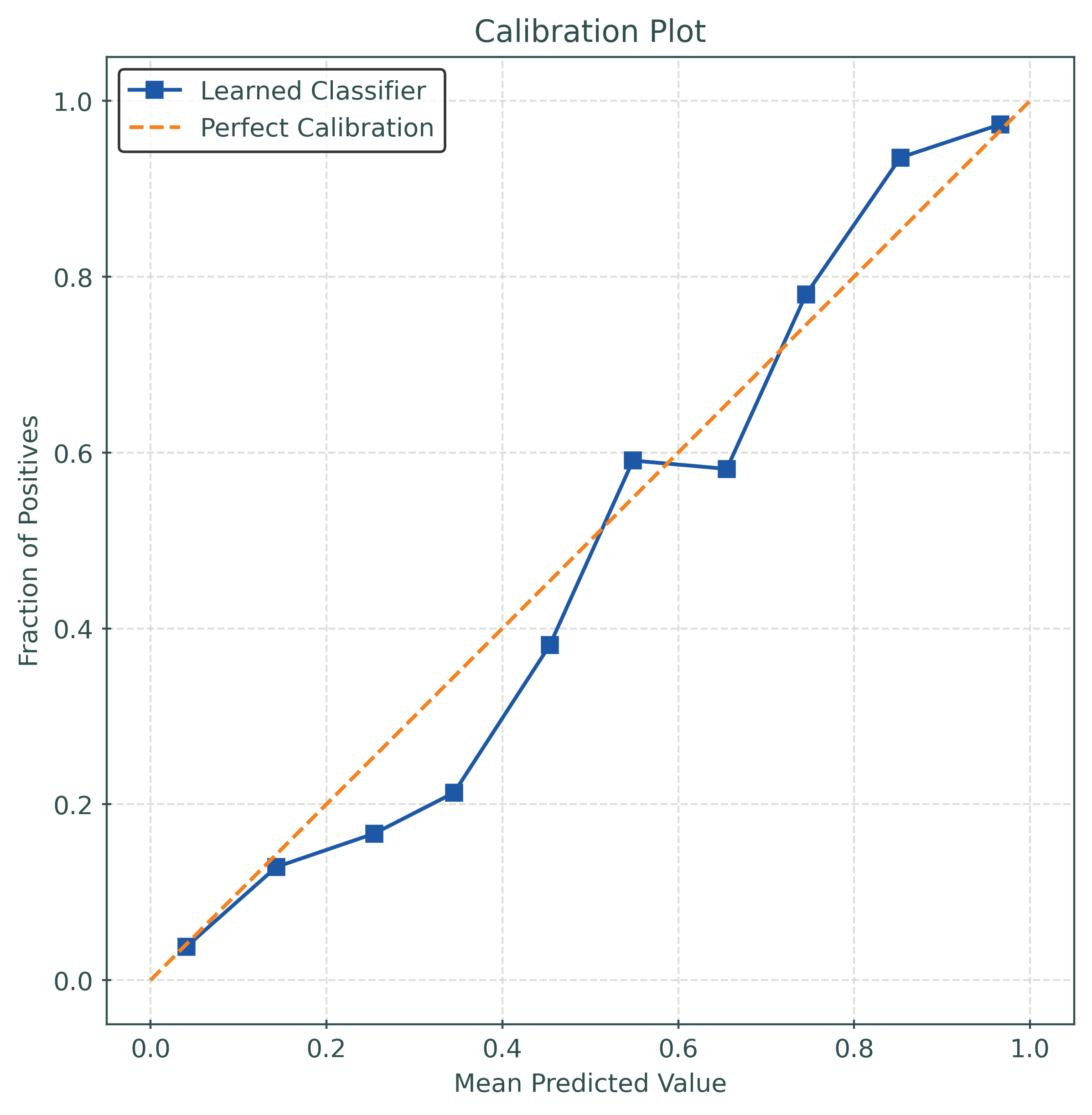
print(f"Test Accuracy: {test_accuracy:.4f}")
print(f"Log Loss: {test_logloss:.4f}")
print(f"Brier Score: {test_brier_score:.4f}")Test Accuracy: 0.8770
Log Loss: 0.3116
Brier Score: 0.0941# function to calculate the calibration error
def calibration_error(y_true, y_prob, type="expected", n_bins=10):
"""
Compute calibration error of a binary classifier.
The calibration error measures the aggregated difference between
the average predicted probabilities assigned to the positive class,
and the frequencies of the positive class in the actual outcome.
Parameters
----------
y_true : array-like of shape (n_samples,)
True targets of a binary classification task.
y_prob : array-like of (n_samples,)
Estimated probabilities for the positive class.
type : {'expected', 'max'}, default='expected'
The expected-type is the Expected Calibration Error (ECE), and the
max-type corresponds to Maximum Calibration Error (MCE).
n_bins : int, default=10
The number of bins used when computing the error.
Returns
-------
score : float
The calibration error.
"""
bins = np.linspace(0.0, 1.0, n_bins + 1)
bin_idx = np.searchsorted(bins[1:-1], y_prob)
bin_sums = np.bincount(bin_idx, weights=y_prob, minlength=len(bins))
bin_true = np.bincount(bin_idx, weights=y_true, minlength=len(bins))
bin_total = np.bincount(bin_idx, minlength=len(bins))
nonzero = bin_total != 0
prob_true = bin_true[nonzero] / bin_total[nonzero]
prob_pred = bin_sums[nonzero] / bin_total[nonzero]
if type == "max":
calibration_error = np.max(np.abs(prob_pred - prob_true))
elif type == "expected":
bin_error = np.abs(prob_pred - prob_true) * bin_total[nonzero]
calibration_error = np.sum(bin_error) / len(y_true)
return calibration_errorExpected Calibration Error (ECE)
\[ \text{ECE}(y, p) = \sum_{m=1}^M \frac{|B_m|}{n} | \bar{y}(B_m) - \bar{p}(B_m) | \]
Maximum Calibration Error (MCE)
\[ \text{MCE}(y, \hat{p}) = \max_m | \bar{y}(B_m) - \bar{p}(B_m) | \]
- \(n\), the number of samples
- \(y\), the vector of true labels
- \(p\), the vector of probabilities that \(y_i = 1\) given \(x_i\)
- \(M\), the number of bins
- \(|B_m|\), the number of samples in the \(m\)th bin
- \(\bar{y}(B_m)\), the proportion of samples in the \(m\)th bin with \(y_i = 1\)
- \(\bar{y}(B_m) = \frac{1}{|B_m|} \displaystyle\sum_{i \in B_m} y_i\)
- \(\bar{p}(B_m)\), the average probability in the \(m\)th bin
- \(\bar{p}(B_m) = \frac{1}{|B_m|} \displaystyle\sum_{i \in B_m} p_i\)
test_ece = calibration_error(
y_test,
y_pred_proba,
type="expected",
)
print(f"Test ECE: {test_ece:.4f}")Test ECE: 0.0351# function to plot a calibration_plot
def plot_calibration_plot(y_true, y_prob):
# generate "data" for calibration plot
prob_true, prob_pred = calibration_curve(
y_true,
y_prob,
n_bins=10,
pos_label=1,
)
# create a figure and axis object with a specific size
fig, ax = plt.subplots()
# plot the calibration curve
ax.plot(
prob_pred,
prob_true,
"s-",
label="Learned Classifier",
color="#1D58A7",
)
# plot the diagonal "perfect" line
ax.plot(
[0, 1],
[0, 1],
"--",
label="Perfect Calibration",
color="#F5821E",
)
# set the plot title and axis labels
ax.set_title("Calibration Plot")
ax.set_xlabel("Mean Predicted Value")
ax.set_ylabel("Fraction of Positives")
# add a grid
ax.grid(
True,
color="lightgrey",
linewidth=0.75,
linestyle="--",
)
# fix aspect ratio
ax.set_aspect(
"equal",
adjustable="box",
)
# show the legend
ax.legend()
# show the plot
plt.show()# create a calibrated model
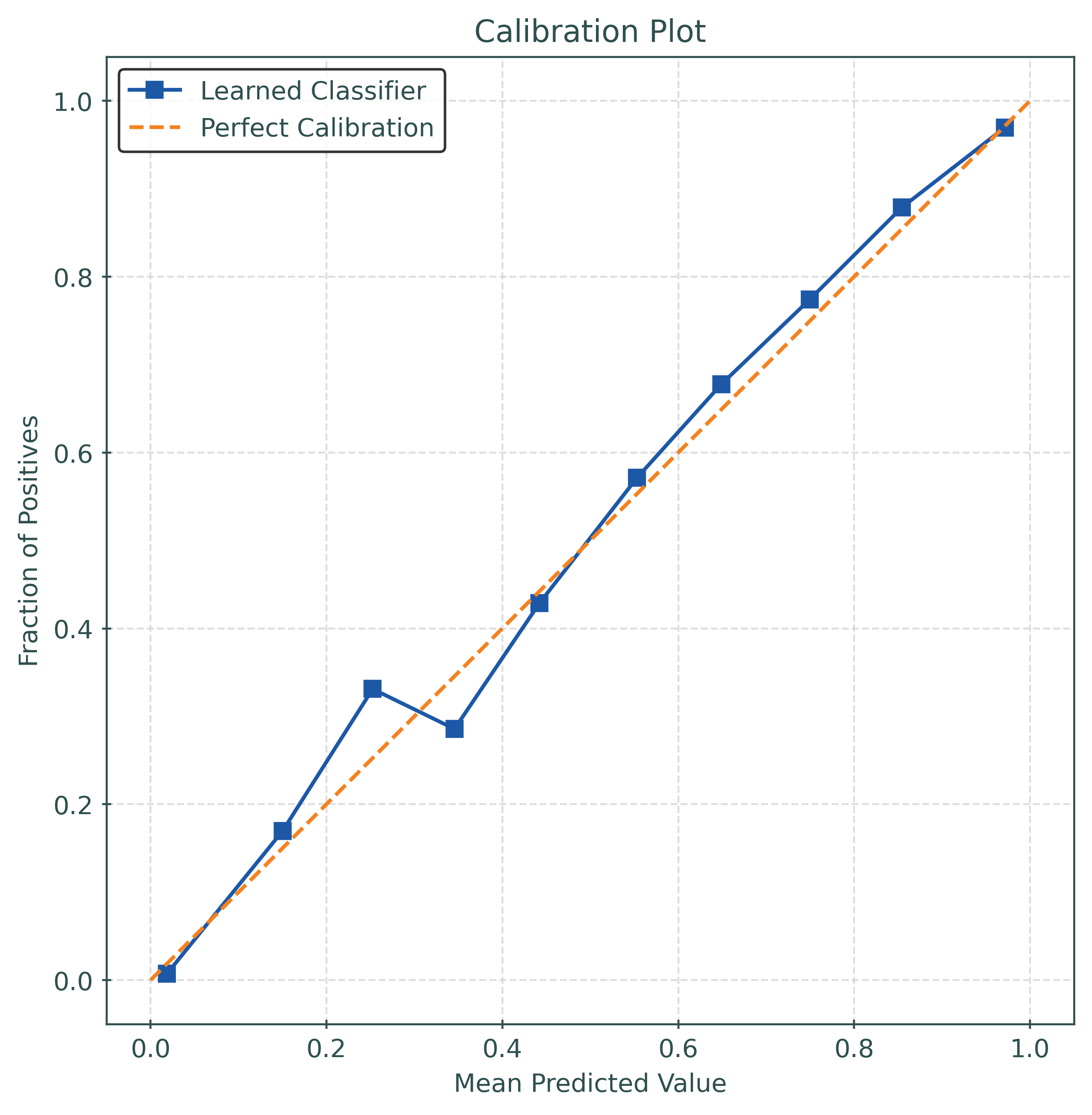
model_calibrated = CalibratedClassifierCV(
estimator=LogisticRegression(random_state=42),
method="isotonic",
)
model_calibrated.fit(X_train, y_train)CalibratedClassifierCV(estimator=LogisticRegression(random_state=42),
method='isotonic')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
CalibratedClassifierCV(estimator=LogisticRegression(random_state=42),
method='isotonic')LogisticRegression(random_state=42)
LogisticRegression(random_state=42)
# get predicted probabilities and classification
y_pred_proba_calibrated = model_calibrated.predict_proba(X_test)[:, 1]
y_pred_calibrated = model_calibrated.predict(X_test)# calculate metrics
test_accuracy_calibrated = accuracy_score(y_test, y_pred_calibrated)
test_logloss_calibrated = log_loss(y_test, y_pred_proba_calibrated)
test_brier_score_calibrated = brier_score_loss(y_test, y_pred_proba_calibrated)
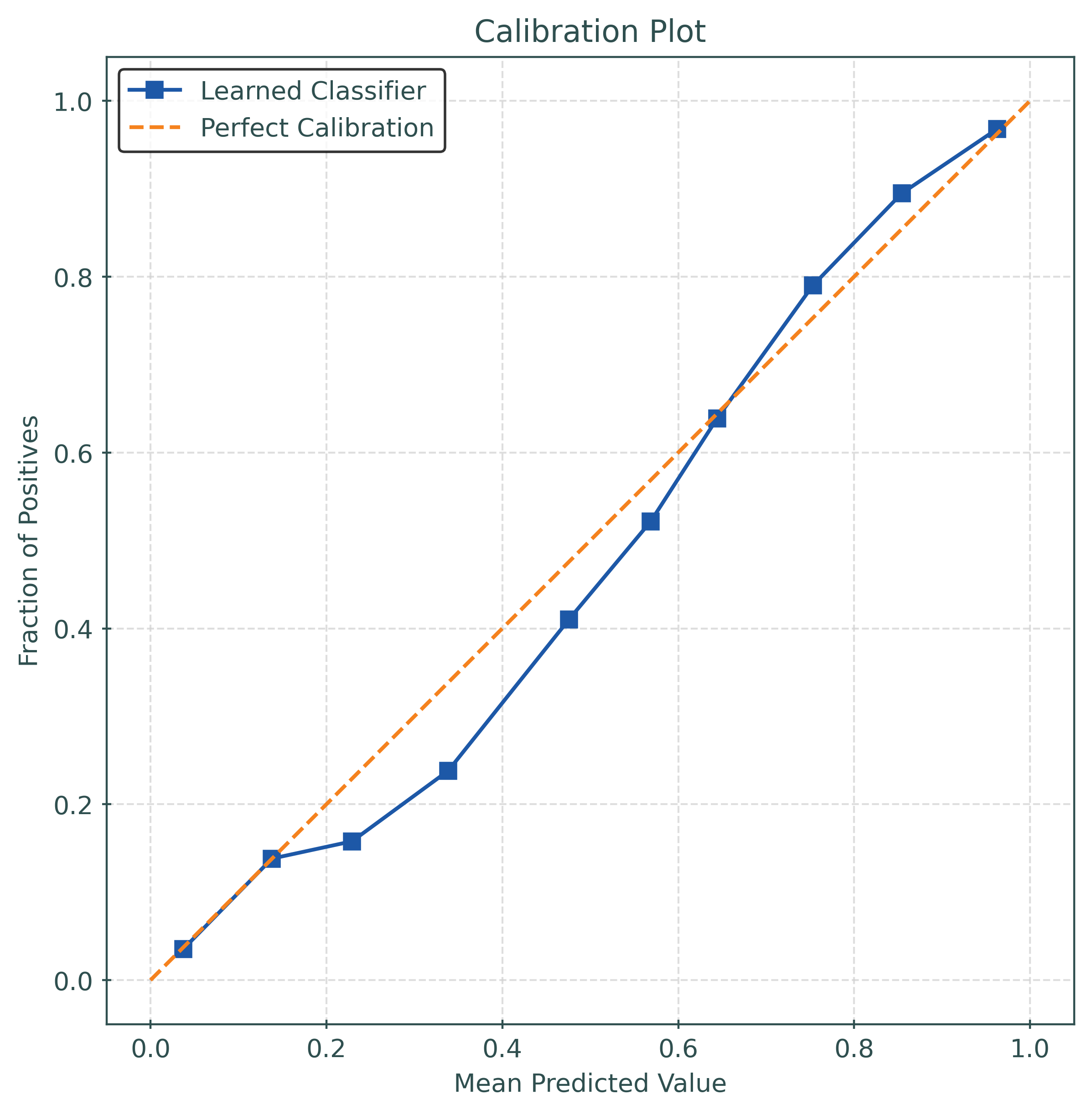
print(f"Calibrated Test Accuracy: {test_accuracy_calibrated:.4f}")
print(f"Calibrated Log Loss: {test_logloss_calibrated:.4f}")
print(f"Calibrated Brier Score: {test_brier_score_calibrated:.4f}")Calibrated Test Accuracy: 0.8780
Calibrated Log Loss: 0.3077
Calibrated Brier Score: 0.0932test_ece_calibrated = calibration_error(
y_test,
y_pred_proba_calibrated,
type="expected",
)
print(f"Test ECE: {test_ece_calibrated:.4f}")Test ECE: 0.0208Tuning
X, y = make_classification(
n_samples=5000,
n_features=20,
n_informative=2,
n_redundant=2,
random_state=42,
)
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.5,
random_state=42,
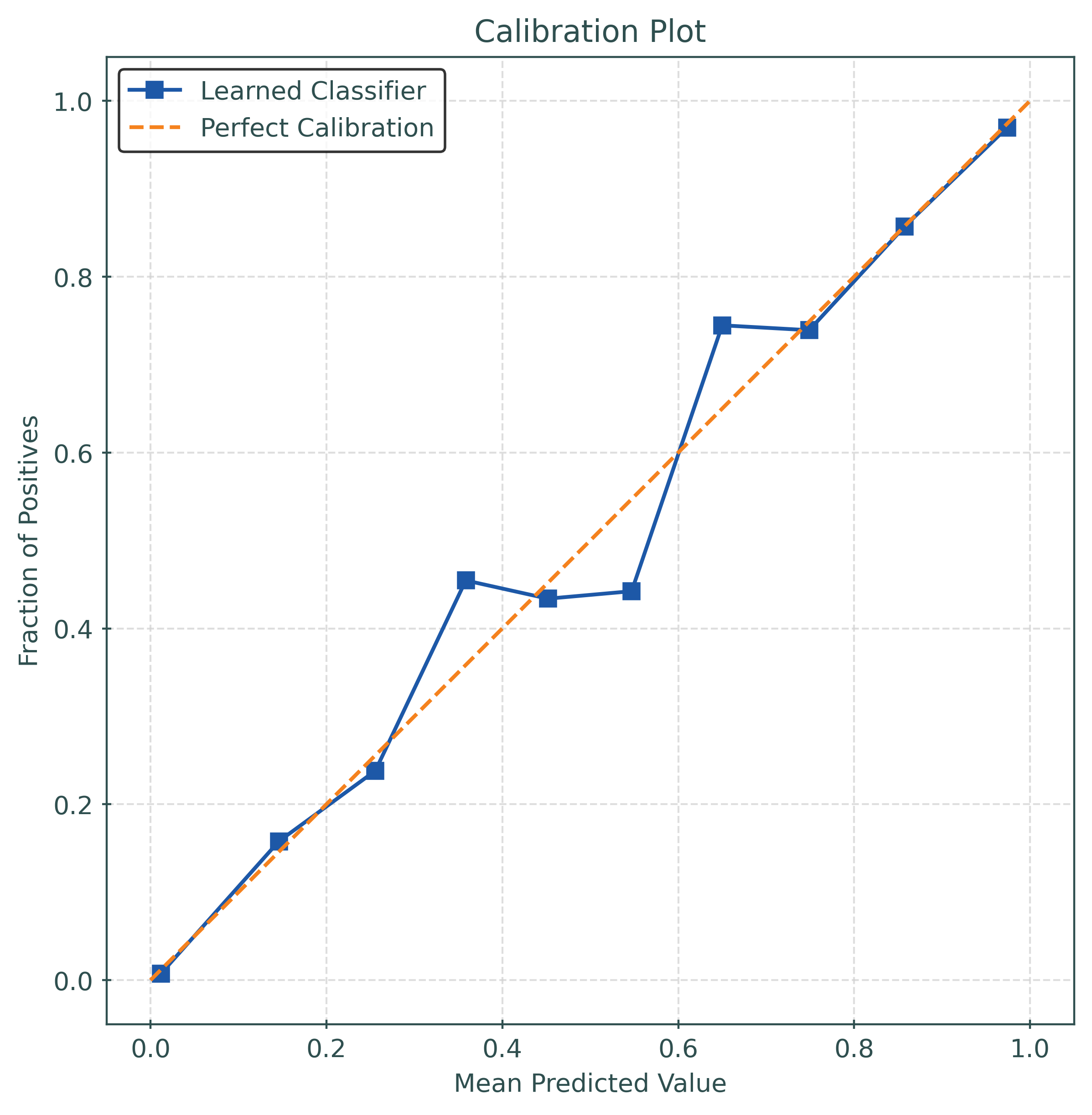
)mod = RandomForestClassifier(
n_estimators=25,
random_state=42,
)mod.fit(X_train, y_train)
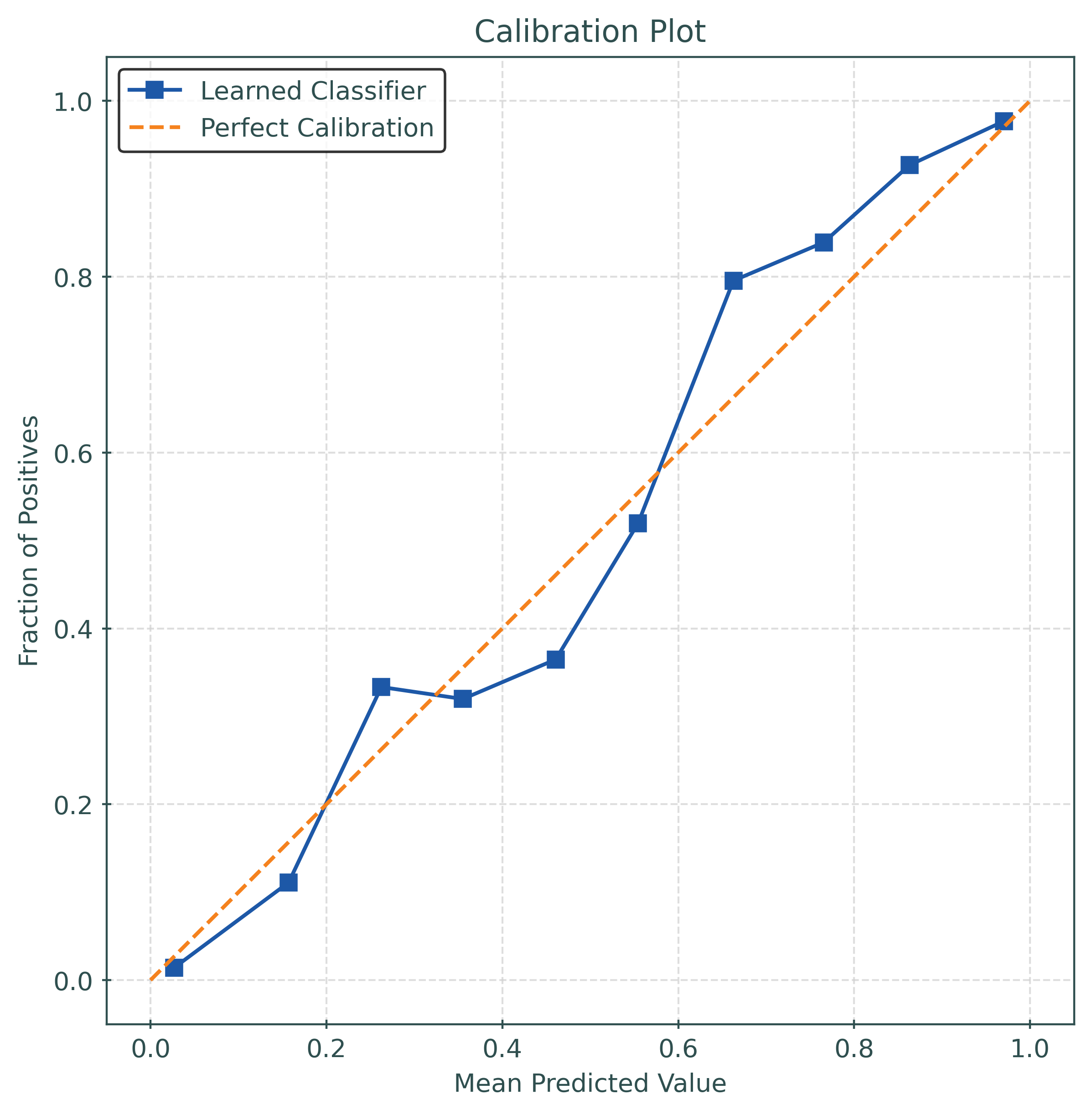
y_pred_proba = mod.predict_proba(X_test)[:, 1]calibration_error(
y_test,
y_pred_proba,
type="expected",
)0.02916799999999984mod = CalibratedClassifierCV(
estimator=RandomForestClassifier(
n_estimators=25,
random_state=42,
),
method="sigmoid",
)mod.fit(X_train, y_train)
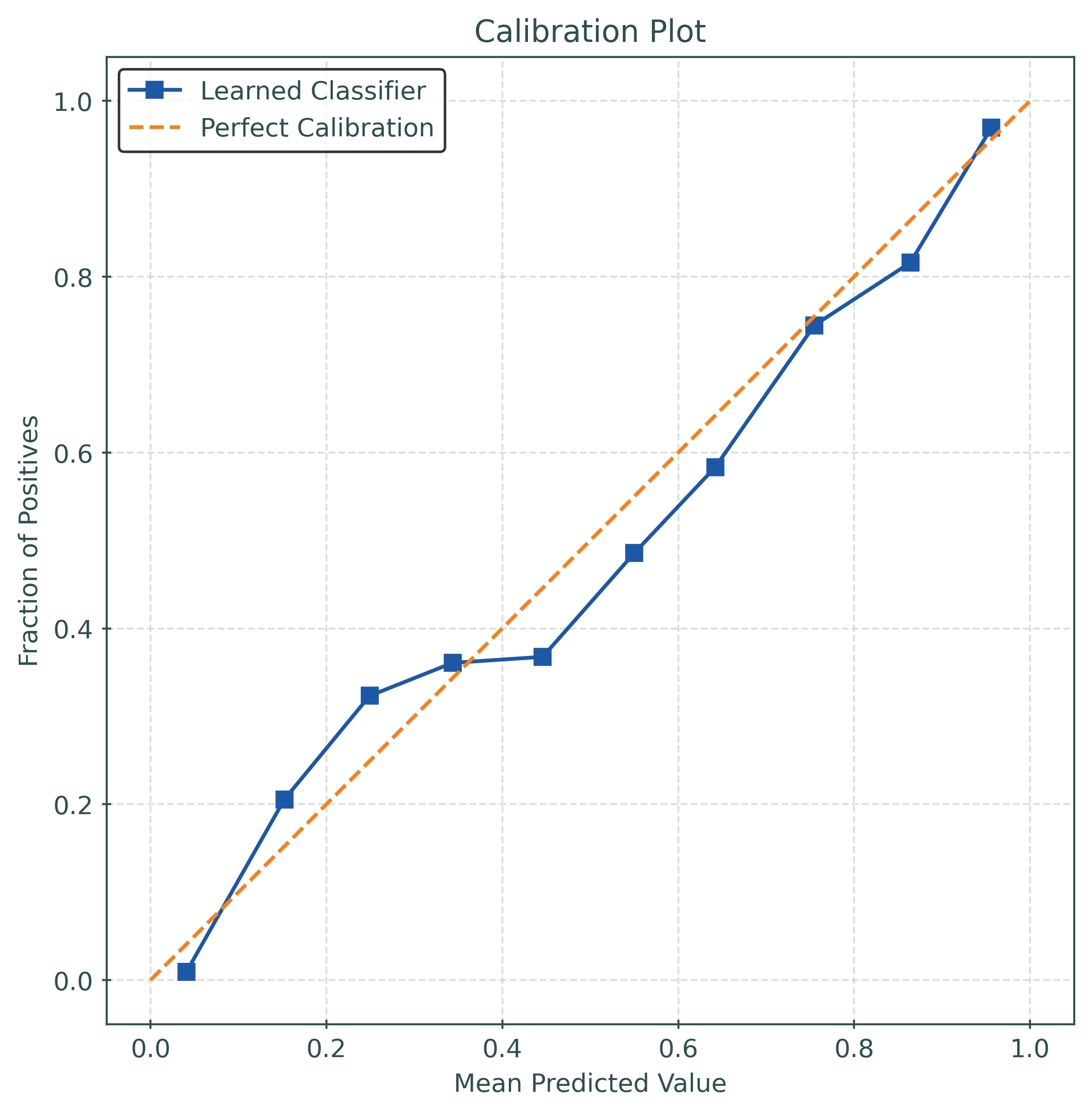
y_pred_proba = mod.predict_proba(X_test)[:, 1]calibration_error(
y_test,
y_pred_proba,
type="expected",
)0.030140064773678886params_grid = {
"method": [
"isotonic",
"sigmoid",
],
"estimator__max_depth": [5, 10, 15, 25, None],
"estimator__criterion": ["log_loss", "gini"],
}mod = GridSearchCV(
CalibratedClassifierCV(
estimator=RandomForestClassifier(
n_estimators=25,
random_state=42,
),
),
cv=5,
param_grid=params_grid,
n_jobs=-1,
)mod.fit(X_train, y_train)
y_pred_proba = mod.predict_proba(X_test)[:, 1]calibration_error(
y_test,
y_pred_proba,
type="expected",
)0.016471495846085135mod = GridSearchCV(
CalibratedClassifierCV(
estimator=RandomForestClassifier(
n_estimators=25,
random_state=42,
),
),
cv=5,
param_grid=params_grid,
n_jobs=-1,
scoring="neg_brier_score",
)mod.fit(X_train, y_train)
y_pred_proba = mod.predict_proba(X_test)[:, 1]calibration_error(
y_test,
y_pred_proba,
type="expected",
)0.016707691547294713