# basic imports
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# model imports
from sklearn.neighbors import KNeighborsRegressor
from sklearn.neighbors import KNeighborsClassifier
# metric imports
from sklearn.metrics import root_mean_squared_error
from sklearn.metrics import accuracy_score
# model selection imports
from sklearn.model_selection import train_test_split
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
# preprocessing imports
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.compose import make_column_transformer
from sklearn.pipeline import make_pipeline
# data imports
from palmerpenguins import load_penguinsCross-Validation
Tuning with Resampling
Validation and Test Data
Before introducing cross-validation, you might still be wondering: Why do we need both a validation set and a test set? Why not just a test set? That certainly would be easier!
- The validation set is used to select a model, through hyperparameter tuning.
- The test set is used to evaluate the model.
But again, why can’t the test set be used to select and evaluate a model? Aren’t we evaluating the model when we select it? Indeed we are, but that evaluation is only used for selection. The evaluation performed using the test data is the final evaluation of the model, which gives an estimate of how the model will perform on new, unseen data.
When we use the validation data to select a model, we are “seeing” or “looking” at that data, despite not fitting the model with that data.
Using only a test set for both selection and evaluation would lead to an optimistic estimate of the model’s performance.
The following simulations studies demonstrate the optimistic bias that results from using the test set for both selection and evaluation.
def simulate_sin_data(n, sd, seed):
np.random.seed(seed)
X = np.random.uniform(
low=-2 * np.pi,
high=2 * np.pi,
size=(n, 1),
)
signal = np.sin(X).ravel()
noise = np.random.normal(
loc=0,
scale=sd,
size=n,
)
y = signal + noise
return X, yCorrect Approach: Validation and Test Data
rmse_tests = []
rmse_trues = []
k_selected_validation = []
for sim in range(250):
X, y = simulate_sin_data(
n=500,
sd=0.25,
seed=sim,
)
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.20,
random_state=42,
)
X_vtrain, X_validation, y_vtrain, y_validation = train_test_split(
X_train,
y_train,
test_size=0.20,
random_state=42,
)
rmse_validations = []
k_values = np.arange(1, 20, 2)
for k in k_values:
knn = KNeighborsRegressor(n_neighbors=k)
knn.fit(X_vtrain, y_vtrain)
validation_rmse = root_mean_squared_error(
y_validation,
knn.predict(X_validation),
)
rmse_validations.append(validation_rmse)
best_k = k_values[np.argmin(rmse_validations)]
k_selected_validation.append(best_k)
knn = KNeighborsRegressor(n_neighbors=best_k)
knn.fit(X_train, y_train)
test_rmse = root_mean_squared_error(y_test, knn.predict(X_test))
rmse_tests.append(test_rmse)
X_new, y_new = simulate_sin_data(
n=10000,
sd=0.25,
seed=42,
)
rmse_true = root_mean_squared_error(y_new, knn.predict(X_new))
rmse_trues.append(rmse_true)
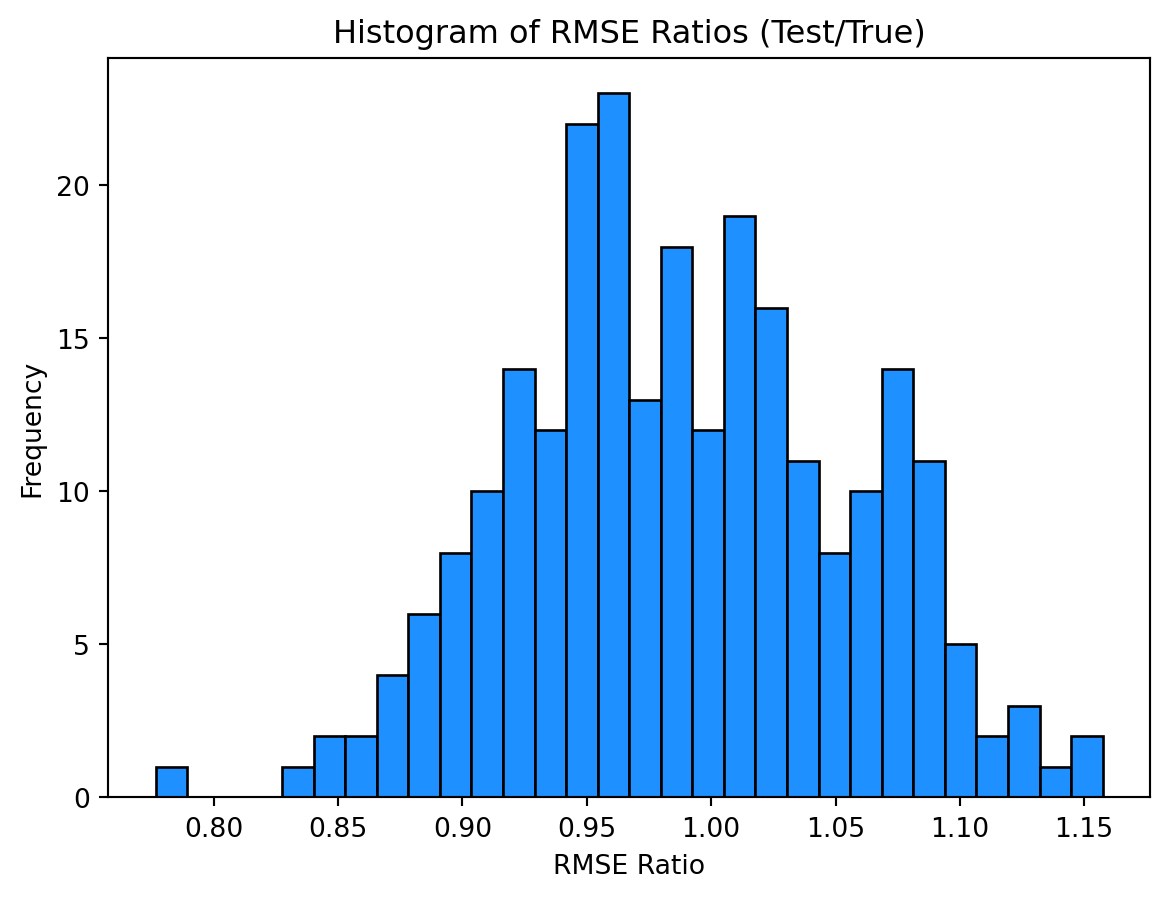
print(np.mean(np.array(rmse_tests) / np.array(rmse_trues)))
print(np.mean(np.array(rmse_tests) / np.array(rmse_trues) < 1))0.9887479393487324
0.58Optimistic Approach: Test Data Only
rmse_tests = []
rmse_trues = []
k_selected_test = []
for sim in range(250):
X, y = simulate_sin_data(
n=500,
sd=0.25,
seed=sim,
)
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.20,
random_state=42,
)
rmse_validations = []
k_values = np.arange(1, 20, 2)
for k in k_values:
knn = KNeighborsRegressor(n_neighbors=k)
knn.fit(X_train, y_train)
validation_rmse = root_mean_squared_error(
y_test,
knn.predict(X_test),
)
rmse_validations.append(validation_rmse)
best_k = k_values[np.argmin(rmse_validations)]
k_selected_test.append(best_k)
knn = KNeighborsRegressor(n_neighbors=best_k)
knn.fit(X_train, y_train)
test_rmse = root_mean_squared_error(y_test, knn.predict(X_test))
rmse_tests.append(test_rmse)
X_new, y_new = simulate_sin_data(
n=10000,
sd=0.25,
seed=42,
)
rmse_true = root_mean_squared_error(y_new, knn.predict(X_new))
rmse_trues.append(rmse_true)
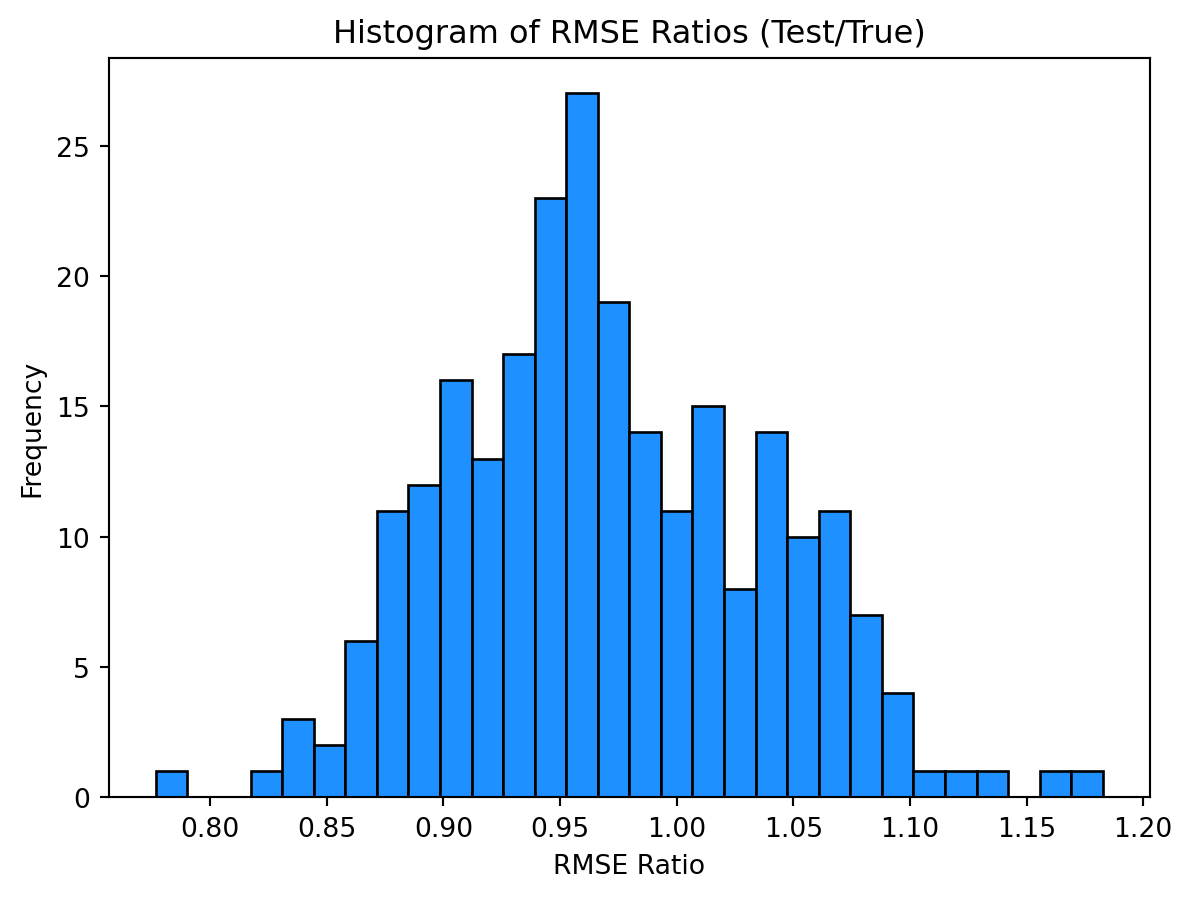
print(np.mean(np.array(rmse_tests) / np.array(rmse_trues)))
print(np.mean(np.array(rmse_tests) / np.array(rmse_trues) < 1))0.9702873893641341
0.688Cross-Validation
Now that we understand the need for both a validation and test set, we will further complicate the supervised learning process by introducing cross-validation. We’ll first introduce cross-validation, then explain the issue that it solves.
Let’s show an example of calculating 5-fold cross-validated RMSE for a K-Nearest Neighbors regression model with \(k=3\).
What is 5-fold cross-validation? 5-fold cross-validation is a specific case of \(k\)-fold cross-validation, where \(k=5\). Essentially, we will split the data into 5 equal parts called “folds.” Each fold will be used as a validation set while the remaining folds are used as the training set, essentially repeating our previous validation process five times. Each time through, we will calculate a metric, for example RMSE. The final cross-validated RMSE will be the average of the five RMSE values.
First, we’ll get some data and train-test split it. Then we’ll perform 5-fold cross-validation on the training data.
X, y = simulate_sin_data(
n=20,
sd=0.25,
seed=sim,
)X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.20,
random_state=42,
)kf = KFold(n_splits=5, shuffle=True, random_state=42)
knn = KNeighborsRegressor(n_neighbors=3)rmse_folds = []
for i, (vtrain_idx, validation_idx) in enumerate(kf.split(X_train)):
print(f" Validation Set: Fold {i} --------------------------------")
print(f" Train Indices: {vtrain_idx}")
print(f" Validation Indices: {validation_idx}")
knn.fit(X_train[vtrain_idx, :], y_train[vtrain_idx])
y_pred = knn.predict(X_train[validation_idx, :])
rmse_fold = root_mean_squared_error(y_train[validation_idx], y_pred)
print(f" Validation RMSE: {rmse_fold:.3f}.")
print(f"")
rmse_folds.append(rmse_fold)
print(f"5-Fold Cross-Validated RMSE: {np.mean(rmse_folds):.3f}") Validation Set: Fold 0 --------------------------------
Train Indices: [ 2 3 4 6 7 8 9 10 11 12 13 15]
Validation Indices: [ 0 1 5 14]
Validation RMSE: 0.708.
Validation Set: Fold 1 --------------------------------
Train Indices: [ 0 1 2 3 4 5 6 7 9 10 12 14 15]
Validation Indices: [ 8 11 13]
Validation RMSE: 0.448.
Validation Set: Fold 2 --------------------------------
Train Indices: [ 0 1 3 4 5 6 7 8 10 11 12 13 14]
Validation Indices: [ 2 9 15]
Validation RMSE: 0.403.
Validation Set: Fold 3 --------------------------------
Train Indices: [ 0 1 2 3 5 6 8 9 11 12 13 14 15]
Validation Indices: [ 4 7 10]
Validation RMSE: 0.863.
Validation Set: Fold 4 --------------------------------
Train Indices: [ 0 1 2 4 5 7 8 9 10 11 13 14 15]
Validation Indices: [ 3 6 12]
Validation RMSE: 0.249.
5-Fold Cross-Validated RMSE: 0.534Why would we need to do this? Doesn’t this just make things more complicated? Doesn’t this just waste a bunch of time? We’re just repeating the same thing five times!
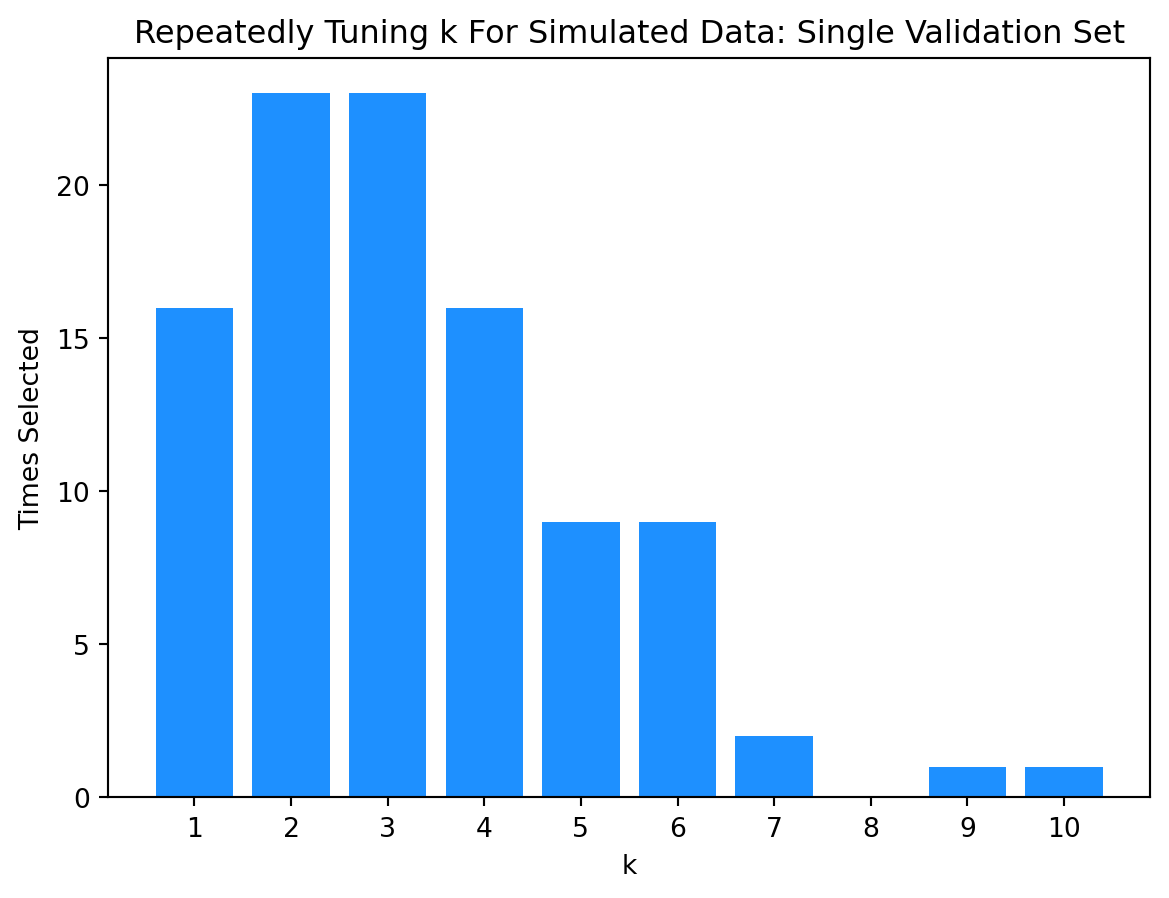
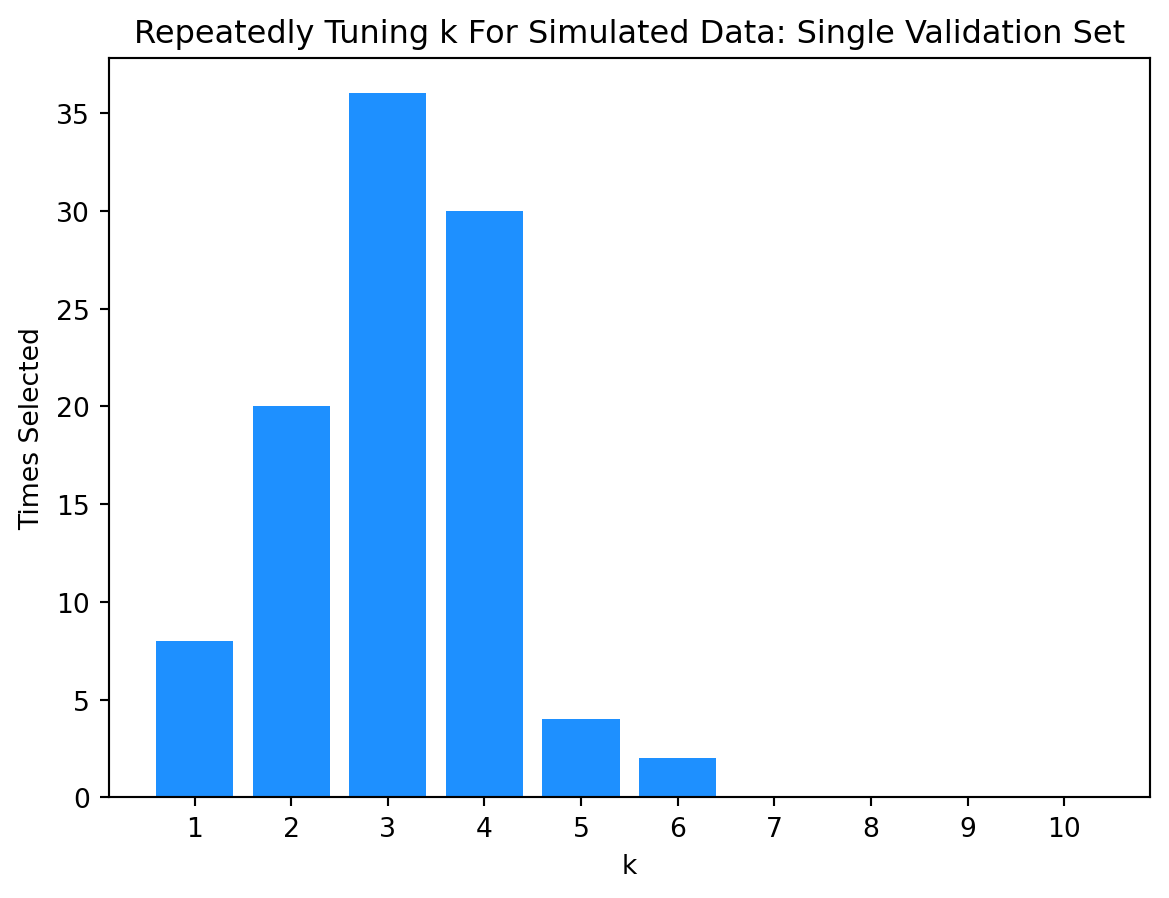
Let’s perform another simulation study. This time we’ll compare the results of selecting a model with a single validation set versus selecting a model with 5-fold cross-validation.
# here we define the values of k we will consider
k_values = np.arange(1, 11)Tuning with Single Validation Set
k_best = []
for seed in range(100):
X, y = simulate_sin_data(
n=50,
sd=0.25,
seed=sim,
)
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.20,
random_state=seed,
)
X_vtrain, X_validation, y_vtrain, y_validation = train_test_split(
X_train,
y_train,
test_size=0.20,
random_state=seed,
)
k_rmse = []
for k in k_values:
knn = KNeighborsRegressor(n_neighbors=k)
knn.fit(X_vtrain, y_vtrain)
k_rmse.append(root_mean_squared_error(y_validation, knn.predict(X_validation)))
k_best.append(k_values[np.argmin(k_rmse)])np.array(k_best)array([ 6, 6, 6, 2, 5, 2, 6, 2, 9, 4, 1, 2, 6, 7, 2, 3, 3,
1, 1, 3, 1, 2, 3, 3, 2, 3, 2, 1, 2, 3, 5, 2, 2, 1,
5, 4, 5, 2, 4, 2, 2, 5, 2, 10, 5, 1, 7, 3, 3, 4, 3,
2, 6, 1, 6, 1, 2, 4, 4, 1, 1, 4, 4, 6, 2, 4, 2, 4,
4, 3, 3, 3, 3, 4, 1, 3, 5, 2, 4, 2, 3, 3, 2, 6, 3,
1, 5, 4, 3, 4, 4, 3, 2, 5, 1, 1, 3, 3, 1, 3])Tuning with Cross-Validation
k_best = []
for seed in range(100):
X, y = simulate_sin_data(
n=50,
sd=0.25,
seed=sim,
)
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.20,
random_state=seed,
)
k_rmse = []
for k in k_values:
knn = KNeighborsRegressor(n_neighbors=k)
cv_scores = cross_val_score(
knn,
X_train,
y_train,
cv=5,
scoring="neg_root_mean_squared_error",
)
k_rmse.append(-np.mean(cv_scores))
k_best.append(k_values[np.argmin(k_rmse)])np.array(k_best)array([4, 1, 4, 6, 3, 3, 5, 4, 2, 3, 4, 2, 4, 4, 4, 2, 3, 4, 2, 3, 2, 2,
2, 3, 3, 3, 4, 1, 4, 2, 4, 4, 3, 1, 4, 3, 2, 3, 4, 4, 4, 3, 5, 4,
3, 4, 4, 1, 3, 2, 3, 2, 5, 3, 4, 3, 4, 3, 4, 2, 1, 2, 3, 4, 3, 4,
3, 3, 3, 3, 3, 3, 4, 6, 2, 2, 3, 3, 3, 1, 3, 3, 3, 3, 2, 5, 2, 4,
4, 3, 4, 4, 2, 2, 1, 3, 4, 3, 1, 2])Grid Search
So far, we’ve demonstrated two things:
- The need for both validation and test data.
- The need for cross-validation.
But while we’ve shown that we need to do these things, we haven’t shown how to do them efficiently. The code we’ve written so far is not the code you would write in practice.
Instead, in practice you will want to use the GridSearchCV class, together with a Pipeline. Combining these two tools will demonstrate the true power of sklearn. At times you might need something more custom, or an alternative to GridSearchCV, but for most cases, GridSearchCV and a Pipeline will be your go-to when using sklearn to perform supervised learning.
By using GridSearchCV and a Pipeline, rather than writing for loops and other verbose code to perform parameter tuning, instead you can focus on specifying the inputs to the process. As the human in the loop, you will need to specify:
- The model(s) you want to tune.
- The hyperparameters you want to tune.
- The values you want to consider for each hyperparameter.
- The metric you want to use to evaluate the models.
- The number of folds for cross-validation.
- The preprocessing steps you want to include in the pipeline.
Let’s take a look at an example.
Example: Palmer Penguins
penguins = load_penguins()penguins_train, penguins_test = train_test_split(
penguins,
test_size=0.35,
random_state=42,
)penguins_train| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year | |
|---|---|---|---|---|---|---|---|---|
| 292 | Chinstrap | Dream | 50.3 | 20.0 | 197.0 | 3300.0 | male | 2007 |
| 302 | Chinstrap | Dream | 50.5 | 18.4 | 200.0 | 3400.0 | female | 2008 |
| 56 | Adelie | Biscoe | 39.0 | 17.5 | 186.0 | 3550.0 | female | 2008 |
| 271 | Gentoo | Biscoe | NaN | NaN | NaN | NaN | NaN | 2009 |
| 10 | Adelie | Torgersen | 37.8 | 17.1 | 186.0 | 3300.0 | NaN | 2007 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 188 | Gentoo | Biscoe | 42.6 | 13.7 | 213.0 | 4950.0 | female | 2008 |
| 71 | Adelie | Torgersen | 39.7 | 18.4 | 190.0 | 3900.0 | male | 2008 |
| 106 | Adelie | Biscoe | 38.6 | 17.2 | 199.0 | 3750.0 | female | 2009 |
| 270 | Gentoo | Biscoe | 47.2 | 13.7 | 214.0 | 4925.0 | female | 2009 |
| 102 | Adelie | Biscoe | 37.7 | 16.0 | 183.0 | 3075.0 | female | 2009 |
223 rows × 8 columns
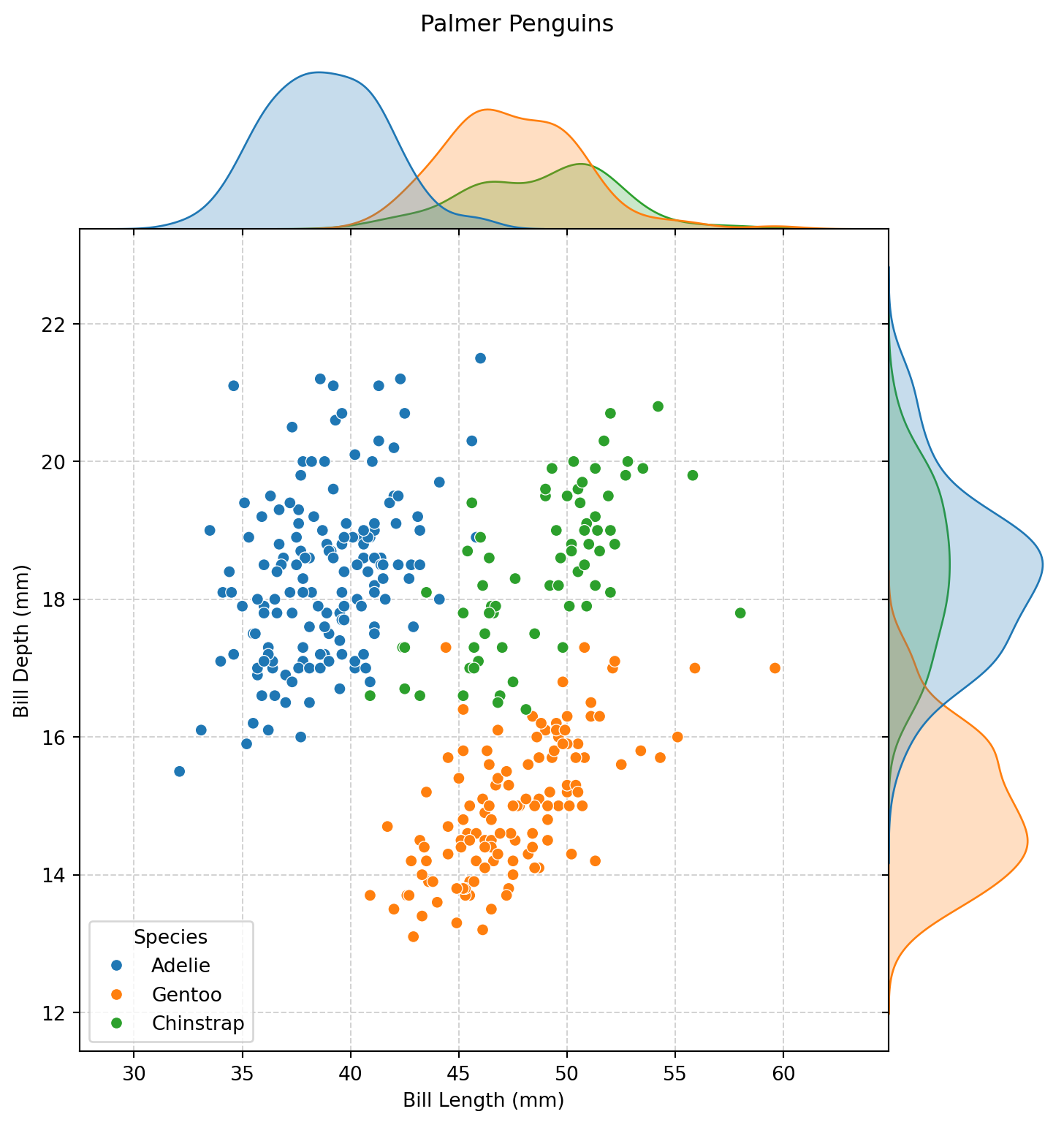
plot = sns.jointplot(
data=penguins,
x="bill_length_mm",
y="bill_depth_mm",
hue="species",
space=0,
zorder=2,
)
plot.set_axis_labels(
xlabel="Bill Length (mm)",
ylabel="Bill Depth (mm)",
)
plot.figure.suptitle(
t="Palmer Penguins",
y=1.02,
)
plot.ax_joint.legend(
title="Species",
loc="lower left",
)
plot.ax_joint.grid(
color="lightgrey",
linestyle="--",
linewidth=0.75,
zorder=1,
)
plot.figure.set_size_inches(
w=8,
h=8,
)numeric_features = [
"bill_length_mm",
"bill_depth_mm",
"body_mass_g",
]
categorical_features = ["sex"]
features = numeric_features + categorical_features
target = "species"X_train = penguins_train[features]
y_train = penguins_train["species"]
X_test = penguins_test[features]
y_test = penguins_test["species"]# define preprocessing for numeric features
numeric_transformer = make_pipeline(
SimpleImputer(strategy="median"),
StandardScaler(),
)
# define preprocessing for categorical features
categorical_transformer = make_pipeline(
SimpleImputer(strategy="most_frequent"),
OneHotEncoder(handle_unknown="infrequent_if_exist"),
)
# create general preprocessor
preprocessor = make_column_transformer(
(numeric_transformer, numeric_features),
(categorical_transformer, categorical_features),
remainder="drop",
)
# define the classifier
knn = KNeighborsClassifier()
# define the parameter grid
param_grid = {
"kneighborsclassifier__n_neighbors": [1, 5, 10, 15, 20, 25, 50],
"kneighborsclassifier__p": [1, 2],
}
# create the pipeline
pipeline = make_pipeline(
preprocessor,
knn,
)
# create the grid search
mod = GridSearchCV(
pipeline,
param_grid,
cv=5,
scoring="accuracy",
)
# fit the grid search
mod.fit(X_train, y_train)
# print the best parameters and the corresponding score
print(f"Best Parameters: {mod.best_params_}")
print(f"Best Cross-Validated Accuracy: {mod.best_score_}")Best Parameters: {'kneighborsclassifier__n_neighbors': 1, 'kneighborsclassifier__p': 1}
Best Cross-Validated Accuracy: 0.9821212121212122# create a DataFrame to store the CV results
cv_results = pd.DataFrame(mod.cv_results_)
# select relevant columns and sort by rank
cv_results = cv_results[
[
"param_kneighborsclassifier__n_neighbors",
"param_kneighborsclassifier__p",
"mean_test_score",
"std_test_score",
"rank_test_score",
]
].sort_values(by="rank_test_score")
# rename columns for better readability
cv_results.columns = [
"n_neighbors",
"p",
"mean_test_score",
"std_test_score",
"rank_test_score",
]
# display the DataFrame
cv_results| n_neighbors | p | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|
| 0 | 1 | 1 | 0.982121 | 0.016658 | 1 |
| 5 | 10 | 2 | 0.982020 | 0.016955 | 2 |
| 1 | 1 | 2 | 0.977576 | 0.020105 | 3 |
| 2 | 5 | 1 | 0.977576 | 0.028271 | 3 |
| 3 | 5 | 2 | 0.977576 | 0.020105 | 3 |
| 7 | 15 | 2 | 0.977475 | 0.024898 | 6 |
| 11 | 25 | 2 | 0.973030 | 0.016822 | 7 |
| 9 | 20 | 2 | 0.972929 | 0.022311 | 8 |
| 4 | 10 | 1 | 0.968485 | 0.023102 | 9 |
| 6 | 15 | 1 | 0.968485 | 0.023102 | 9 |
| 8 | 20 | 1 | 0.959596 | 0.017014 | 11 |
| 10 | 25 | 1 | 0.941818 | 0.030120 | 12 |
| 13 | 50 | 2 | 0.927879 | 0.044139 | 13 |
| 12 | 50 | 1 | 0.834141 | 0.017144 | 14 |
# predict on the test data
y_test_pred = mod.predict(X_test)
# calculate and print the test accuracy
test_accuracy = accuracy_score(y_test, y_test_pred)
print(f"Test Accuracy: {test_accuracy}")Test Accuracy: 0.9834710743801653